Multitasking Neural Networks for Multiplanar MRI Prostate Localization and Segmentation
By Hosseiny M, Masoudi S, Conlin C, Zhong A, Retson T, Bahrami N, You S, Hussain T, Digma L, Babar HS, Seibert T, Hsiao A
Abstract
Purpose: Deep learning can be a powerful tool for automating visual tasks in medical imaging, including the localization and segmentation of anatomic structures. However, training these algorithms often requires substantial data curation that may require expert image annotation on multiple imaging planes. For example, the base and apex of the prostate may be more readily marked by radiologists in sagittal planes, while segmentation of central and peripheral zones of the prostate may be more readily handled in axial planes. We thus sought to develop a deep-learning (DL) strategy capable of integrating annotations across multiple imaging planes and hypothesized that it would outperform traditional algorithms developed using single-plane imaging data only.
Materials and Methods: In this retrospective, IRB-approved, HIPAA-compliant study, we collected pelvic magnetic resonance images (MRIs) from 656 male patients. The urinary bladder, prostate, and point locations of the prostate’s apex and base were annotated on sagittal T2 images in 391 patients. Central and peripheral zones of the prostate were segmented on axial T2 images in 265 patients. Datasets were then divided by patient into training (80%), validation (10%), and test (10%) cohorts.
Three convolutional neural networks (CNNs) were trained, each based on a U-Net architecture: CNN1 using sagittal images to provide heatmap localizations of the apex and base of the prostate, CNN2 using axial images to segment the prostate and divide it into central and peripheral zones, and a multitask CNN using both data sets to accomplish both tasks. To this end, images and annotations were transformed into a common coordinate system and a custom conditional loss function was incorporated to handle missing labels and encode three-dimensional geometric relationships. Model performance for segmentation and localization was assessed by Dice score and L2 distance error.
Results: Median Dice for whole prostate segmentation improved from 0.803 (IQR 0.76-0.818) by CNN1 to 0.882 (IQR, 0.842-0.890) by the multitask CNN (p < 0.05, Wilcoxon test). Mean dice scores for central and peripheral zone segmentation were [81.02 ± 5.58, 63.36 ± 3.96%] respectively by CNN2 and [81.02 ± 5.58 %, 61.67 ± 7.04%] by the multitask CNN respectively without significant difference. Median L2 errors for localization of the base and apex of the prostate decreased from 5.7 (IQR 4.5-6.9) mm and 6.5 (IQR 5.6-7.7) mm by CNN1 to 3.6 (IQR 2.6-4.7) mm and 3.5 (IQR 2.4-4.5) mm by the multitask CNN.
Conclusions: Our proposed multi-task CNN was capable of learning both segmentation and localization tasks, incorporating data from multiple imaging planes, and exceeding the performance of the CNNs trained on individual tasks. These results demonstrate the potential of CNNs for tackling related visual tasks and their potential for combining data from multiple sources or imaging planes.
Keywords: Multitask, Deep learning, Prostate, MRI
Introduction
Medical image segmentation involves partitioning an input image into different segments and aims to delineate the foreground anatomical or pathological structures from the background. Image segmentation helps in the analysis of medical images by highlighting regions of interest, which can be used to define anatomic boundaries, for detection of abnormalities in computer-assisted diagnosis, dose planning for radiotherapy, surgery simulation, and other forms of treatment decision-making. 1
Manual segmentation by a human expert might seem like the simplest solution to define target boundaries; however, it is a time-consuming and user-dependent process.2 Because it is such a foundational aspect of so many biomedical problems, segmentation continues to be an important area of ongoing research.3 Deep learning algorithms, and specifically convolutional neural networks (CNNs), are capable of learning and reproducing an extensive range of parameters, which can then be used to extract features from medical images. 4,5
In the last several years, deep learning has emerged as a powerful tool for automating segmentation of anatomic structures in medical images. Although each CNN algorithm is generally developed to accomplish a single task, multitask CNNs have the potential to be more computationally efficient. The potential performance benefit of multitask CNNs remains unclear, though few studies have shown improved performance of multi-task algorithms for segmentation and classification of breast tumors on ultrasound and mammography. 6,7
Image segmentation for prostate MRI can be utilized for a variety of aspects of medical care, including delineation of the gland and zonal boundaries for measurements of prostatic enlargement, procedural planning for prostate biopsies, and radiation therapy planning. 8-10 In addition, algorithms that locate the base and apex of the prostate gland can help to delineate the prostate gland’s spatial orientation for automating MRI scan prescription 11,12 and standardizing 3D reconstructions.
However, as with many tasks in medical imaging, certain anatomic structures are better delineated in one plane than another, partly due to anisotropic spatial resolution of multiplanar MRI. For example, the apex and base of the prostate may be more readily delineated in the sagittal plane, and the zonal boundaries of the prostate may be more readily delineated in the axial plane. It remains unclear how to best combine data from multiple imaging planes, and to what degree combining such information is beneficial for CNN performance.
We thus sought to explore the potential of a multitask CNN to combine multiplanar MR images and annotations, and to evaluate its performance for accomplishing two tasks: 1) dividing the prostate into central and peripheral zones, leveraging annotations in axial sections, and 2) localizing the base and apex of the prostate, leveraging annotations in sagittal sections. To that end, we proposed a strategy in which two datasets were aggregated by transforming images and annotations into a common coordinate system and applying a conditional loss function to address missing labels and encode geometric relationships between anatomic structures. We hypothesized that this multitask approach would outperform single models separately trained for individual tasks.
Methods
In this retrospective, IRB-approved, HIPAA-compliant study, we collected a convenience sample of pelvic MRIs from 656 male patients (mean age 67 years, range 38-87). Pelvic MRIs were acquired as part of routine clinical care for initial detection, treatment planning, active surveillance of prostate cancer, or to assess for recurrence in previously treated patients with elevated prostate-specific antigen (PSA).
Image Data
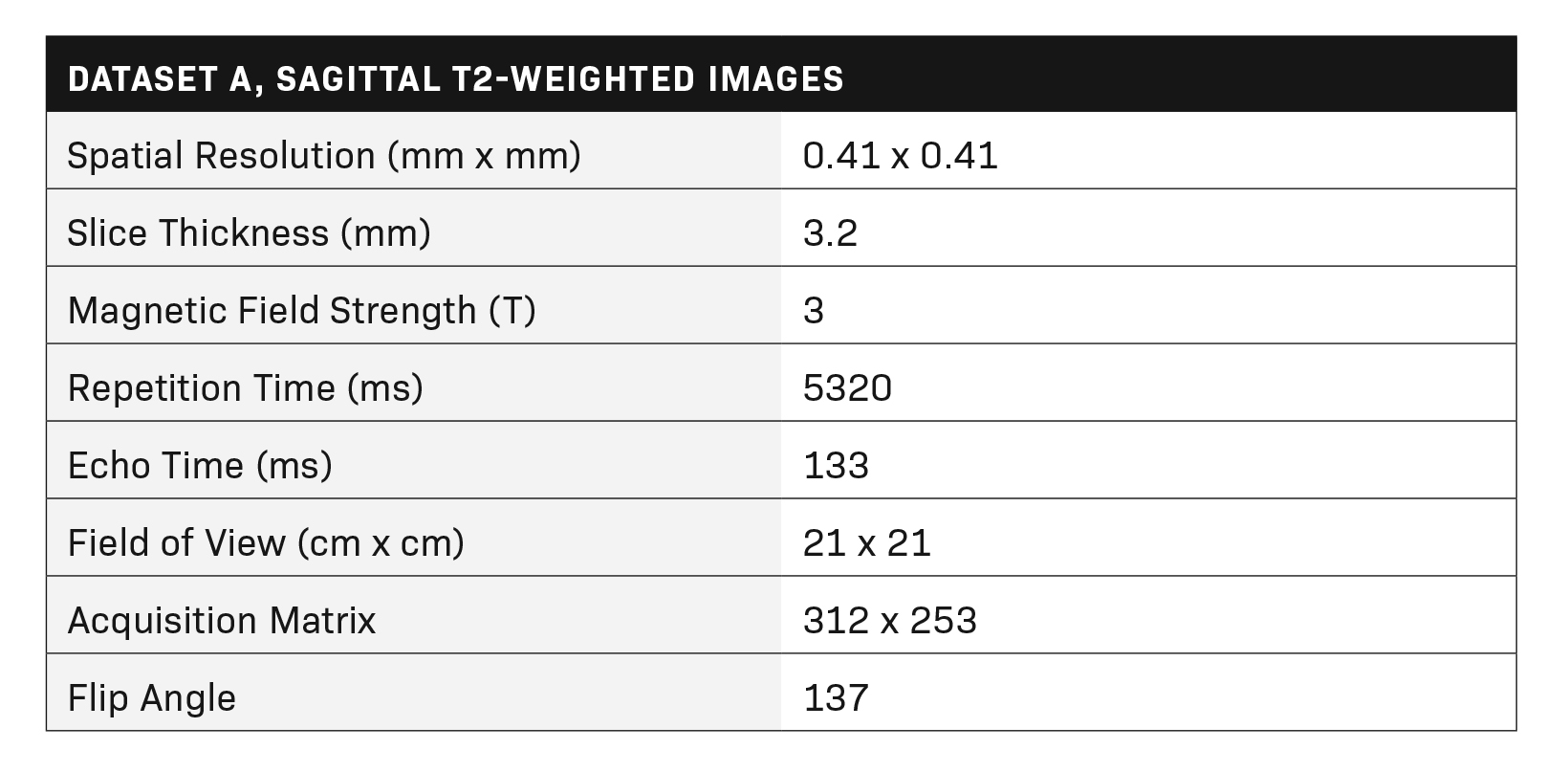
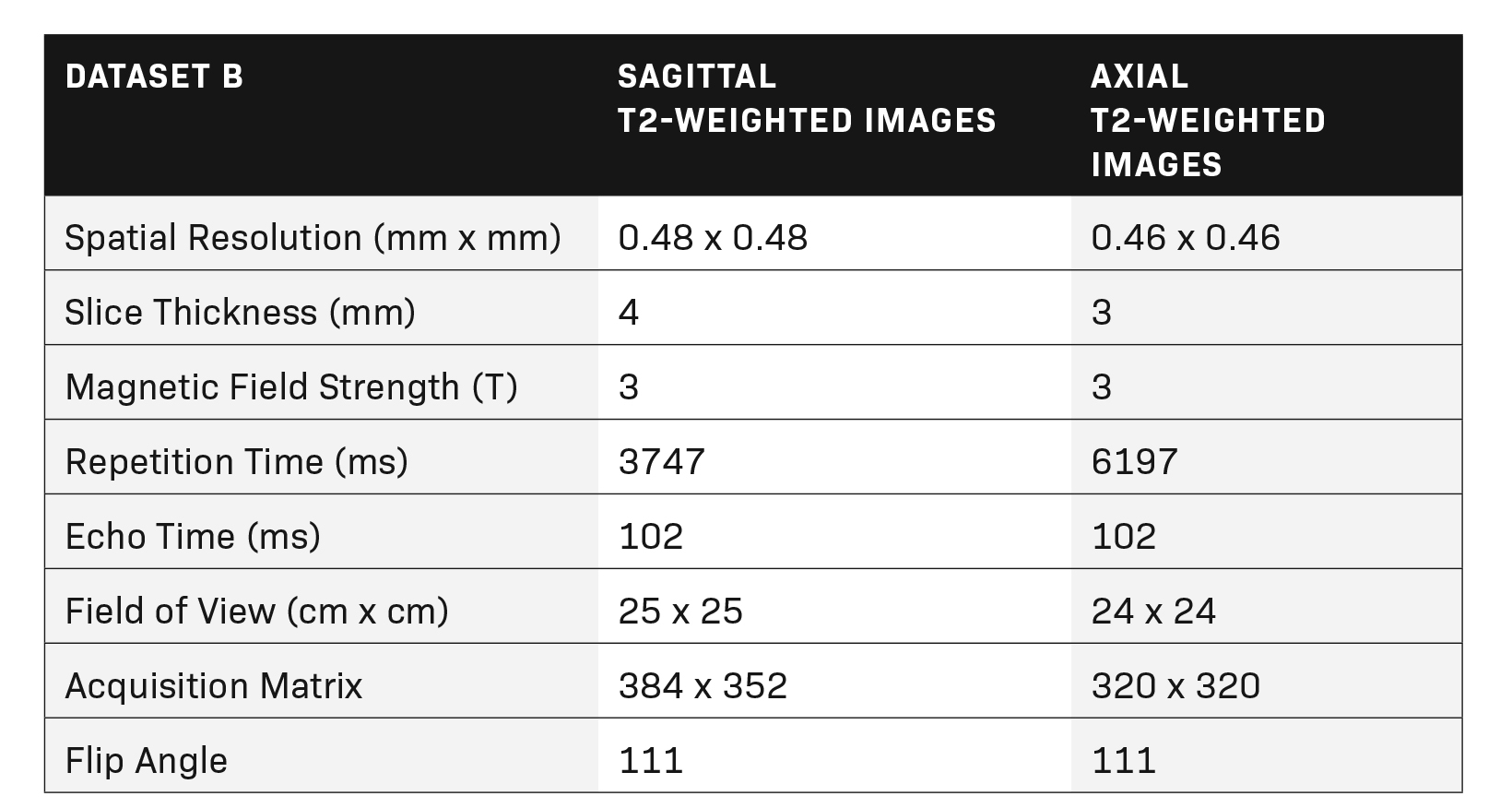
Multiparametric MR imaging of the prostate was performed utilizing an MR scanner with a pelvic external phased-array coil using the same standard protocol in accordance with recommendations of ACR and ESUR. The protocol included two-dimensional turbo spin echo (TSE) T2 imaging, three-dimensional dynamic contrast-enhanced (DCE) imaging in three planes, and echo-planar diffusion-weighted imaging (DWI). Tables 1 and 2 show the MRI acquisition parameters.
Image Annotation
Data for this study comprised two datasets. Dataset A included segmentations of the urinary bladder and prostate, and point localizations of the apex and base of the prostate, each annotated on T2 sagittal images from 391 patients. These annotations were performed by a medical student and a radiology resident, supervised by a board-certified radiologist using Arterys (Tempus, USA) software.
Annotations of the apex and base of the prostate were transformed from point localizations to Gaussian heatmaps. Dataset B included segmentations of the prostate gland and its peripheral zone, each annotated on T2 axial images from 265 patients. These annotations were performed by an image analyst and radiology postdoctoral fellow, supervised by a board-certified radiation oncologist. Each dataset was then divided by patient into training (80%), validation (10%), and test (10%) cohorts.
Model Training
Three modified 3D U-Nets were developed with multiple output channels for segmentation and localization tasks. CNN1 was trained using dataset A and a weighted sum of the segmentation and localization loss functions. CNN2 was trained using dataset B and the segmentation loss only. Axial annotations from dataset B were then translated into their sagittal equivalent and combined with dataset A; this combined dataset was used to train the multitask CNN, incorporating a custom conditional loss function for segmentation and localization in order to 1) ignore missing annotations in the combined dataset, and 2) to take advantage of the morphological relations among the regions of interest. Boundary constraints included the following: (a) central and peripheral zones must sum up to the complete prostate gland; (b) urinary bladder and prostate segmentations must have no overlap, and (c) prostate and base localizations must overlap with the prostate gland segmentation.
In this work, segmentation loss was defined using the Tversky index while localization loss was based on mean square error. For training the single-task CNNs, input images were resampled and zero-padded to (x, y, z) dimensions of 256 x 256 x 16. These images were preprocessed using histogram matching followed by simple image standardization. Augmentation in the form of random image cropping (by -10 to 10 pixels), shifting (by -20 to 20 pixels), and rotation (by -10 to 10 pixels) were applied during neural network training.
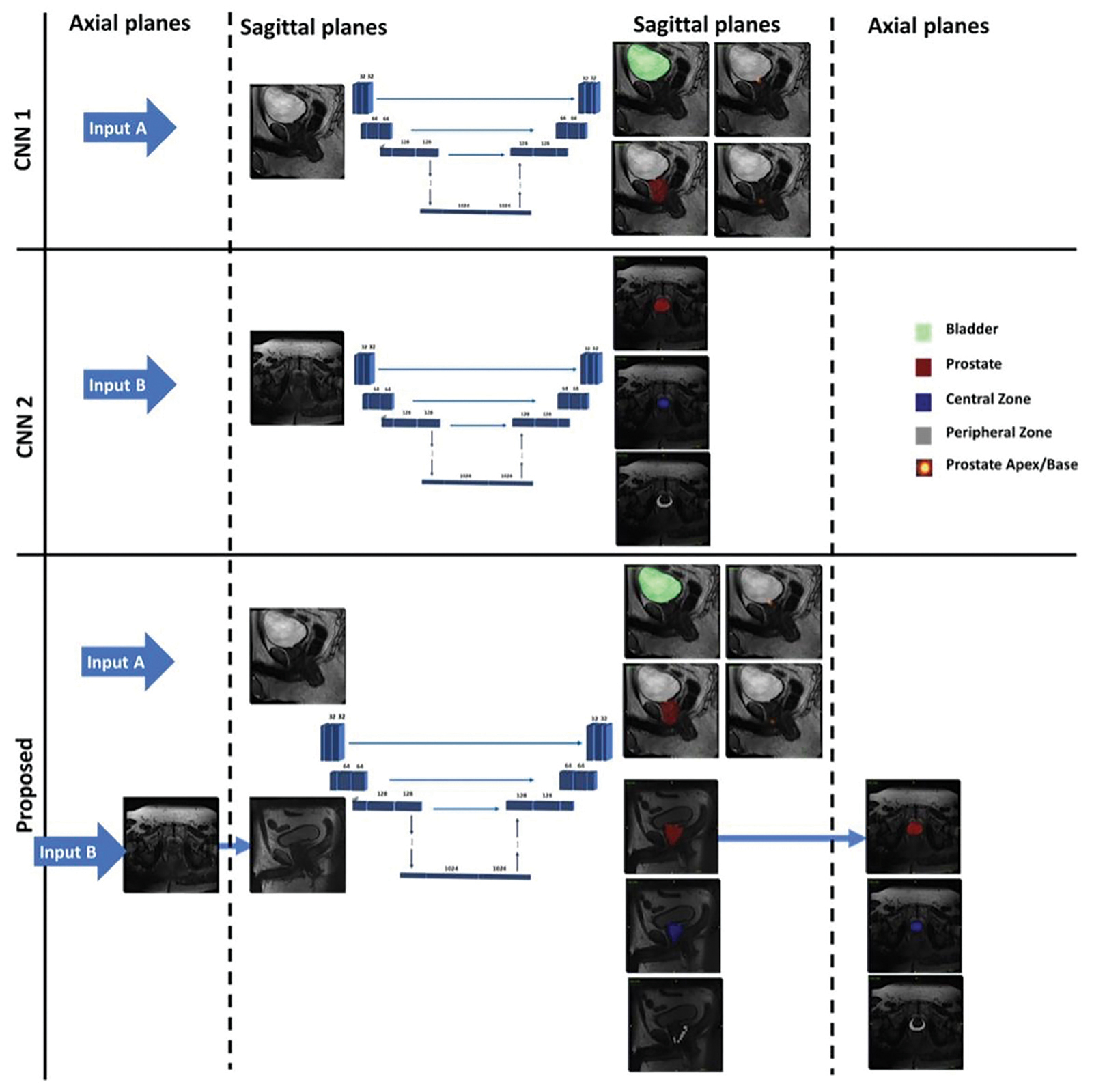
Predictions generated by the multitask CNN were compared directly against CNN1 predictions and translated back to the axial plane for comparison against CNN2. Flowchart of the proposed multi-task CNN and its tasks relative to the single-task CNNs (CNN1 and CNN2) are shown in Figure 1.
Statistical Analysis
Segmentation performance was assessed using Dice scores (expressed as mean ± SD and/or median along with the interquartile range (IQR)). Localization performance was evaluated using landmark L2 distance error and angulation error, calculated as the error between lines connecting the apex and base of the prostate. Wilcoxon tests were used for comparison of Dice scores, L2 distance error, and angulation error between the evaluated models.
Results
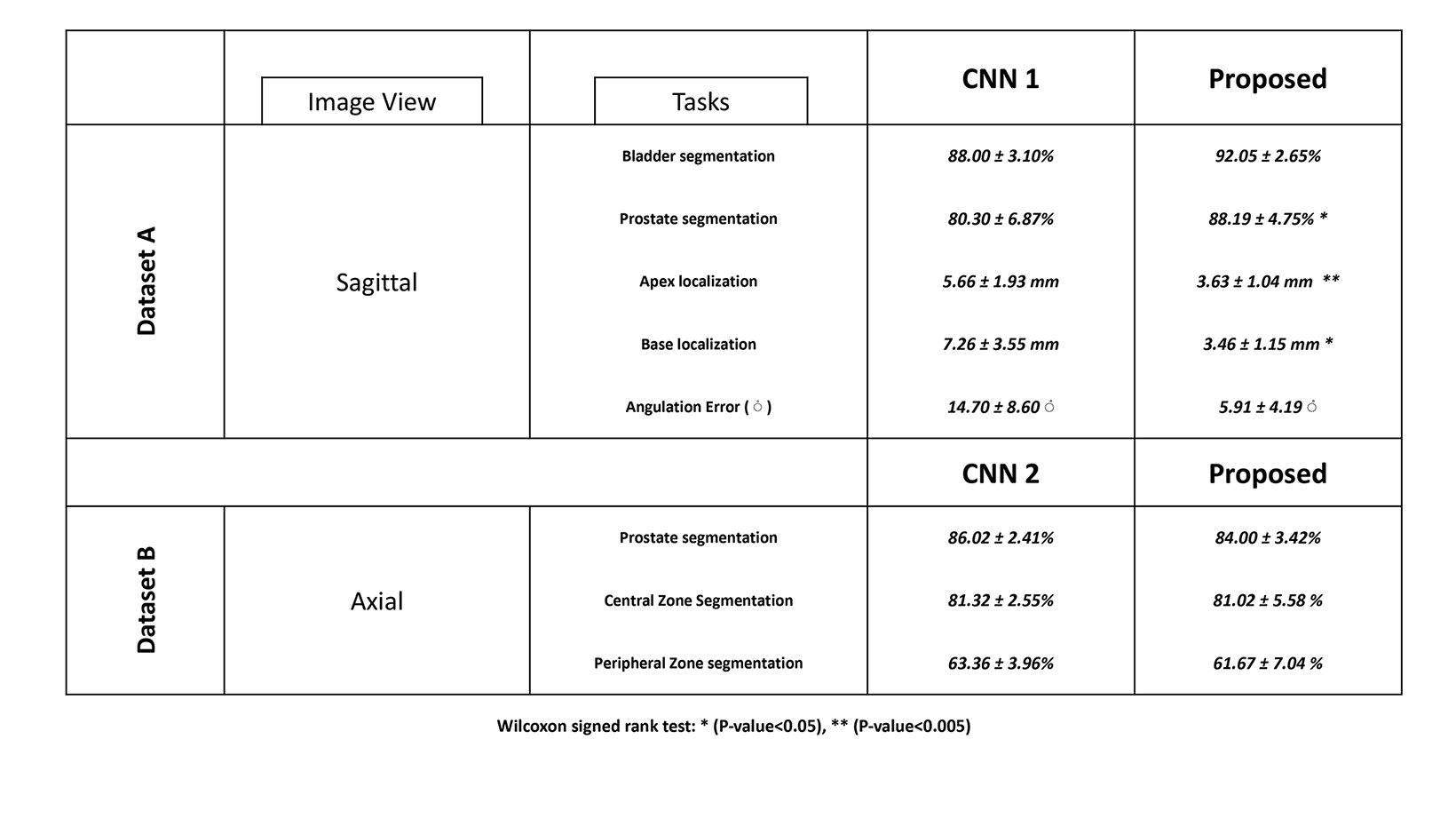
Using two different annotated datasets, three modified 3D U-Nets were trained for segmentation and localization tasks on prostate MRI. Performance of all three CNNs is shown in Table 3.
Prostate Gland Localization
Mean L2 distance error for localization of the prostate gland’s apex and base decreased from 5.7±1.92 mm and 6.5 ±2.5 mm by CNN1 to 3.6 ±1.0 mm and 3.5 ±1.2 mm by the multitask CNN. Mean angulation error decreased from 14.7±8.6° by CNN1 to 5.9±4.2° by the multitask CNN (p-value < 0.05). Median L2 distance error for localization of the apex and base of the prostate decreased from 5.7 mm (IQR 4.5-6.9) and 6.5 mm (IQR 5.6-7.7) by CNN1 to 3.6 mm (IQR 2.6-4.7) and 3.5 mm (IQR 2.4-4.5) by the multitask CNN.
Prostate and Urinary Bladder Segmentation
Mean Dice score for prostate segmentation improved from 80.30 ± 6.87% by CNN1 to 88.19 ± 4.75% by the multitask CNN (p-value < 0.05). Median Dice score for prostate segmentation improved from 80.3% (IQR 76.0 - 81.8%) by CNN1 to 88.2% (IQR 84.2
- 89.0%) by the multitask CNN. Mean Dice score for bladder segmentation increased from 88.00 ± 3.10% by CNN1 to 92.05 ± 2.65% by the multitask CNN (pvalue < 0.01). Median Dice score for bladder segmentation improved from 88.99% (IQR 83.78 - 89.97%) by CNN1 to 91.35% (IQR 89.0-93.2%) by the multitask CNN. Our proposed multitask CNN outperformed the single-task CNNs in all segmentation tasks.
Central and Peripheral Zone Segmentation
For central and peripheral zone segmentation, there were no significant differences between the multitask CNN and CNN2
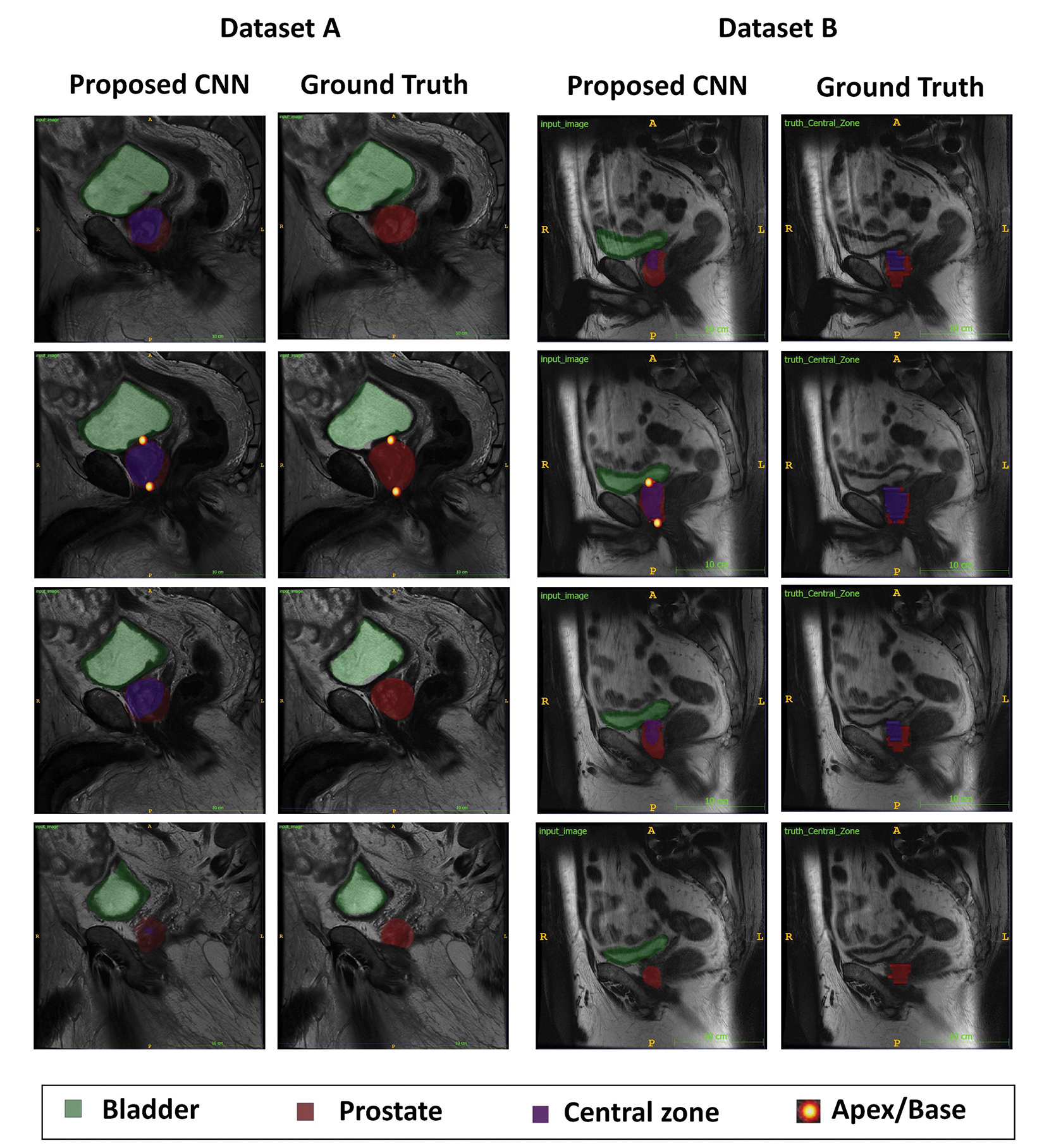
(p-value> 0.05). The mean Dice score for central zone segmentation was 81.02 ± 5.58 % by the multitask CNN and 81.32 ± 2.55% by CNN2. The mean Dice score for peripheral zone segmentation was 61.67 ± 7.04% by the multitask CNN and 63.36 ± 3.96% by CNN2. Two accessions that illustrate multitask CNN performing well are shown in Figure 2.
Discussion
Deep learning is a powerful tool that can be used for the segmentation of anatomic structures in medical imaging, but it typically requires substantial training data that can be time-consuming to obtain. To assess the feasibility of training deep learning algorithms on smaller datasets, we investigated the potential of using multiple datasets, annotated for different purposes on different planes, and aggregated using 3D image re-slicing and image-to-image physical point translation. This is unlike the majority of segmentation approaches presented in the literature which, despite the availability of multi-planar MR images in the standardized protocols, only take axial images into account. The merged dataset was utilized for training a single CNN model, which performed equally well or better than individual models trained using a single dataset.
In the present study, our proposed multitask CNN outperformed the single-task CNNs for pinpointing the apex and base of the prostate, which can be used to define the spatial orientation of the prostate gland. This ultimately can be used for separate ongoing work automating oblique plane prescription in MRI.
The multitask model improved prostate gland and urinary bladder segmentation as well as localization of the prostate base and apex. While the annotations required for each task may be more easily acquired in the axial imaging plane, our proposed CNN model was designed to perform all tasks in a single common coordinate system, utilizing conditional loss functions to address missing labels and encode geometric relationships.
The present study is one of the first to investigate whether a fully automated multitask deep learning algorithm can accomplish multiple tasks while merging two annotated datasets, and if so, how this approach might improve the quality of segmentation and localization results. The combined model yielded significantly decreased mean L2 distances and angulation errors for localization of the prostate gland apex and base by 2.1-3 mm and 8.8°, respectively. Improvement in the automated delineation of the prostate gland’s craniocaudal orientation can potentially enable a faster automated MR imaging prescription. The multi-task model outperformed each single-task model for segmentation of the prostate gland and urinary bladder boundaries by approximately 8% and 4%, respectively, though it did not show a statistically significant difference in zonal segmentation of the prostate gland.
A number of studies have found satisfactory performance of deep learning algorithms for prostate segmentation at ultrasound and mp-MRI. 13,14 In prior investigations with datasets of 49 to 163 patients, CNN models have obtained Dice similarity coefficients ranging from 0.85 to 0.93 for the automatic segmentation of the prostate gland. 15-18 A study by Tian, et al,16 applied a deep-learning algorithm for prostate segmentation on a data set of 140 prostate MRIs and yielded a Dice similarity coefficient of 0.85. The online data collection PROMISE12, which contains labeled prostate MR images, has inspired many studies of prostate segmentations. 19 The present study stands up by employing a larger cohort of training datasets, training a multitask deep learning algorithm, and using multiplanar set of images for performing tasks.
Multi-task training relies on sharing features between related tasks to enable the combined model to perform better on the original single tasks. Training deep learning algorithms using small and partially annotated datasets can also potentially overcome the lack of large training datasets by combining images previously annotated for various purposes on different imaging planes, ultimately facilitating the increasing automation of image analysis tasks.
Limitations
We recognize several limitations to this study. Our proposed multi-task CNN model was trained and validated using retrospective data, so our imaging data includes a variety of different vendors, institutions, and imaging techniques. Further work may be essential to ensure comparable results among other scanner manufacturers and institutional protocols. Failure modes of the multitask CNN model may be revealed through more extensive testing, though additional training data would likely enhance performance. The future direction could focus on how well our proposed multitask CNN performs among patients who have aggressive prostate cancer with invasion to seminal vesicles or bladder. Further investigation is needed to determine how well a multitask CNN model will perform in post-operative patients.
Conclusion
In summary, we show that a multitasking CNN approach can successfully be used to aggregate disparate training data developed for multiple tasks in multiple imaging planes. Multi-task deep learning algorithms that utilize such data can outperform component CNNs trained only on data for individual tasks. We believe a similar approach may be used to perform similar tasks for other organs, paving the way to use datasets of more modest size for image analysis of ever-increasing accuracy and complexity.
References
- Malhotra P, Gupta S, Koundal D, Zaguia A, Enbeyle W. Deep neural networks for medical image segmentation. J Healthc Eng. 2022;2022:9580991. doi:10.1155/2022/9580991
- Comelli A, Dahiya N, Stefano A, et al. Deep learning-based methods for prostate segmentation in magnetic resonance imaging. Appl Sci (Basel). Jan 02 2021;11(2)doi:10.3390/app11020782
- Liu L, Wolterink JM, Brune C, Veldhuis RNJ. Anatomy-aided deep learning for medical image segmentation: a review. Phys Med Biol. 05 26 2021;66(11) doi:10.1088/1361-6560/abfbf4
- Cai L, Gao J, Zhao D. A review of the application of deep learning in medical image classification and segmentation. Ann Transl Med. Jun 2020;8(11):713. doi:10.21037/atm.2020.02.44
- Bardis M, Houshyar R, Chantaduly C, et al. Segmentation of the prostate transition zone and peripheral zone on mr images with deep learning. Radiol Imaging Cancer. 05 2021;3(3):e200024. doi:10.1148/rycan.2021200024
- Zhou Y, Chen H, Li Y, et al. Multi-task learning for segmentation and classification of tumors in 3D automated breast ultrasound images. Med Image Anal. 05 2021;70:101918. doi:10.1016/j.media.2020.101918
- Chowdary J, Yogarajah P, Chaurasia P, Guruviah V. A multi-task learning framework for automated segmentation and classification of breast tumors from ultrasound images. Ultrason Imaging. 01 2022;44(1):3-12. doi:10.1177/01617346221075769
- Tătaru OS, Vartolomei MD, Rassweiler JJ, et al. Artificial intelligence and machine learning in prostate cancer patient management-current trends and future perspectives. Diagnostics (Basel). 02 20 2021;11(2)doi:10.3390/diagnostics11020354
- Hosseiny M, Shakeri S, Felker ER, et al. 3-T Multiparametric MRI followed by in-bore mr-guided biopsy for detecting clinically significant prostate cancer after prior negative transrectal ultrasound-guided biopsy. AJR Am J Roentgenol. 09 2020;215(3):660-666. doi:10.2214/AJR.19.22455
- Hosseiny M, Felker ER, Azadikhah A, et al. Efficacy of 3T multiparametric mr imaging followed by 3t in-bore mr-guided biopsy for detection of clinically significant prostate cancer based on piradsv2.1 score. J Vasc Interv Radiol. Oct 2020;31(10):1619-1626. doi:10.1016/j.jvir.2020.03.002
- Guo Z, Li X, Huang H, Guo N, Li Q. Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans Radiat Plasma Med Sci. Mar 2019;3(2):162-169. doi:10.1109/trpms.2018.2890359
- Masutani EM, Bahrami N, Hsiao A. Deep learning single-frame and multiframe super-resolution for cardiac MRI. Radiology. 06 2020;295(3):552-561. doi:10.1148/radiol.2020192173
- Bardis MD, Houshyar R, Chang PD, et al. Applications of artificial intelligence to prostate multiparametric MRI (mpMRI): current and emerging trends. Cancers (Basel). May 11 2020;12(5)doi:10.3390/cancers12051204
- van Sloun RJG, Wildeboer RR, Mannaerts CK, et al. Deep learning for real-time, automatic, and scanner-adapted prostate (zone) segmentation of transrectal ultrasound, for example, magnetic resonance imaging-transrectal ultrasound fusion prostate biopsy. Eur Urol Focus. 01 2021;7(1):78-85. doi:10.1016/j. euf.2019.04.009
- Clark T, Zhang J, Baig S, Wong A, Haider MA, Khalvati F. Fully automated segmentation of prostate whole gland and transition zone in diffusion-weighted MRI using convolutional neural networks. J Med Imaging (Bellingham). Oct 2017;4(4):041307. doi:10.1117/1.JMI.4.4.041307
- Tian Z, Liu L, Zhang Z, Fei B. PSNet: prostate segmentation on MRI based on a convolutional neural network. J Med Imaging (Bellingham). Apr 2018;5(2):021208. doi:10.1117/1.JMI.5.2.021208
- Karimi D, Samei G, Kesch C, Nir G, Salcudean SE. Prostate segmentation in MRI using a convolutional neural network architecture and training strategy based on statistical shape models. Int J Comput Assist Radiol Surg. Aug 2018;13(8):1211-1219. doi:10.1007/s11548-018-1785-8
- Wang B, Lei Y, Tian S, et al. Deeply supervised 3D fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation. Med Phys. Apr 2019;46(4):1707-1718. doi:10.1002/mp.13416
- Litjens G, Toth R, van de Ven W, et al. Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Med Image Anal. Feb 2014;18(2):359-73. doi:10.1016/j.media.2013.12.002
